Background
I had been working on strand-specific paired-end reads from HiSeq lately and I had trouble mapping reads back to assembled transcripts using Bowtie as well as using RSEM to estimate transcript abundance. After some google searches, I found some information about the dUTP method used to generate stranded reads on Illumina website as well as a blog that explains an orientation of reads from different protocols and which flag (library type) should be used to run Tophat. However, I could not find any information on how to run Bowtie/Bowtie2 as well as RSEM, which uses Bowtie as a default aligner. After a couple days of frustration here is what I learned.
Bowtie and Tophat flags for strand-specific reads
Tophat uses --fr-firststrand for a library created by the dUTP method. This is stated clearly in the manual, so it is easy to understand. In contrast, Bowtie/Bowtie2 uses --fr, --rf, --ff to specify the orientation of paired-end reads.
--fr means the upstream read (/1) is from a forward strand and the downstream read (/2) is from a reverse strand.
--rf means the upstream read (/1) is from a reverse strand and the downstream read (/2) is from a forward strand.
--ff means both reads are from a forward strand.
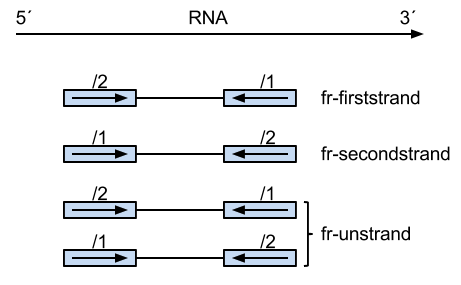
Credit Xianjun Dong.
With --fr flag, the upstream reads could be either /1 and /2, which is valid for unstranded reads. However, as you can see from the figure, the /1 reads from dUTP are from the reverse strand only. So, we need to tell Bowtie not to map the /1 reads to the forward strand. This behavior can be achieved by specifying --nofw flag.
Therefore, to run Bowtie on dUTP reads, the command should look something like this:
bowtie -S -X 1000 --fr --nofw bowtie-index -1 reads_R1.fastq -2 reads_R2.fastq > output.sam
To compare the results, I ran Bowtie/1.0.0 on the same dataset (1M reads) with different flags specified.
| Flag | Mapped Reads | Unmapped Reads |
|---|---|---|
| --fr (default) | 82.57% | 17.43% |
| --rf | 0.17% | 99.83% |
| --ff | 0.00% | 100.00% |
| --fr --nofw | 82.18% | 17.82% |
| --fr --norc | 1.99% | 98.01% |
The number of reads mapped with --fr and --nofw is a bit lower than that from --fr alone. This is due to the fact that without --nofw flag, Bowtie maps /1 reads to both strands.
Note, with --norc flag, Bowtie will not attempt to map the /1 reads to the reverse strand.
Running RSEM on dUTP reads
RSEM has --strand-specific flag that will turn on Bowtie --norc flag, which will force Bowtie to try to map the /1 reads to the forward strand only. This is suitable for reads generated by the ligation method. Also, the --strand-specific flag is equivalent to setting --forward-prob flag to 1.
However, for dUTP reads, the --forward-prob flag should be set to 0 instead. It will specify --nofw flag, which forces Bowtie to map the /1 reads to the reverse strand. Consequently, the --strand-specific flag should not be used when --forward-prob is set to 0. The --forward-prob flag not only specifies --nofw and --norc for Bowtie, but also modifies the orientation parameters of RSEM model.
For example, the command should look something like this:
rsem-calculate-expression -p 8 --forward-prob 0 --paired-end sample_r1.fastq sample_r2.fastq index sample_output
New flags for strand-specific paired-end reads will be added to RSEM in the next minor release. You can read the discussion on this issue here.
Trinity Assembly
To run Trinity with dUTP reads, you need to use RF for --SS_lib_type flag, which means the /1 reads are from the reverse strand and the /2 reads are from the forward strand. Note that, RF flag in Trinity is not the same as --rf in Bowtie.